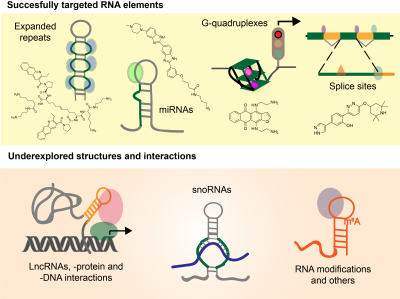
Current Work
Falese, J. P.; McFadden, E. J.; D’Inzeo, C. A.; Hargrove, A. E. “Structural analysis of the lncRNA SChLAP1 reveals protein binding interfaces and a conformationally heterogenous retroviral insertion.” bioRxiv: https://doi.org/10.1101/2021.10.21.465303. In revision.
Wicks, S. L.; Morgan, B. S.; Wilson, A. W.; Hargrove, A. E. “Probing Bioactive Chemical Space to Discover RNA-Targeted Small Molecules.” bioRxiv: https://doi.org/10.1101/2023.07.31.551350\
Bailey, M. A.; Martyr, J. G.; Hargrove, A. E.; Fitzgerald, M. C. “Stability-based Proteomics for Investigation of Structured RNA-protein Interactions.” Anal. Chem. 2024, 96(7), 3044–3053.
https://doi.org/10.1021/acs.analchem.3c04978
Bagnolini, G.; Luu, TB.; Hargrove, AE. “Recognizing the power of machine learning and other computational methods to accelerate progress in small molecule targeting of RNA.” RNA. 2023, 29, 473–488
https://doi.org/10.1261/rna.079497.122
Zafferani, M.; Martyr, J.; Muralidharan, D.; Montalvan, N.; ZhengguoCai; Hargrove, A. E. “Predictive bidirectional modulation of lncRNA MALAT1 triplex stability in vitro through multi-assay profiling of a small molecule library.” ACS Chem. Biol. 2022, 17, 9, 2437–2447
https://doi.org/10.1021/acschembio.2c00124

Donlic, A.; Swanson, E. G.; Chiu, L.-Y.; Wicks, S. L.; Juru, A. U.; Cai, Z.; Kassam, K.; Laudeman, C.; Sanaba, B. G.; Sugarman, A.; Han, E.; Tolbert, B. S.; Hargrove, A. E. “R-BIND 2.0: An Updated Database of Bioactive RNA-Targeting Small Molecules and Associated RNA Secondary Structures.” ACS. Chem. Bio. 2022, 17, 6, 1556-1566 https://doi.org/10.1021/acschembio.2c00224
Cai, Z.; Zafferani, M.; Akande, O.; Hargrove, A. E. “Quantitative Structure Activity Relationship (QSAR) study predicts small molecule binding to RNA structure.” J. Med. Chem. 2022, 65, 10, 7262-7277. https://doi.org/10.1021/acs.jmedchem.2c00254
Zafferani, M.; Muralidharan, D.; Hargrove, A. E. “RT-qPCR as a screening platform for mutational and small molecule impacts on structural stability of RNA tertiary structures.” RSC Chem. Biol. 2022, 3, 905-915. https://doi.org/10.1039/D2CB00015F
Zafferani, M.; Hargrove, A. E. “Small molecule targeting of biologically relevant RNA tertiary and quaternary structures.” Cell Chem. Biol. 2021, 28, 5, 594-609. https://doi.org/10.1016/j.chembiol.2021.03.003
Falese, J. P.; Donlic, A.; Hargrove, A. E. “Tutorial Review: Targeting RNA with small molecules: from fundamental principles towards the clinic.” Chem. Soc. Rev., 2021, 50, 2224–2243. https://doi.org/10.1039/D0CS01261K.
Umuhire Juru, A.; Hargrove, A. E. “Frameworks for targeting RNA with small molecules.” J. Biol. Chem. 2021, 296, 100191. https://doi.org/10.1074/jbc.REV120.015203
Zafferani, M.; Haddad, C.; Luo, L.; Davila-Calderon, J.; Yuan-Chiu, L.; Mugisha, C. S.; Monaghan, A. G.; Kennedy, A. A.; Yesselman, J. D.; Gifford, R. R.; Tai, A. W.; Kutluay, S. B.; Li, M.; Brewer, G.; Tolbert, B. S.; Hargrove, A. E. “Amilorides inhibit SARS-CoV-2 replication in vitro by targeting RNA structures.” Science Advances. 2021, 7. DOI: 10.1126/sciadv.abl6096
This work was featured on Duke’s Research Blog Indy week and the New York Times!
Hargrove, Amanda E. “Small molecule–RNA targeting: starting with the fundamentals.” Chem Comm. 2020, 56, 14744-14756 . DOI: 10.1039/D0CC06796B
This article is part of the themed collection: 2020 Emerging Investigators
Donlic, Anita; Zafferani, Martina; Padroni, Giacomo; Puri, Malavika; Hargrove, Amanda E. “Regulation of MALAT1 triple helix stability and in vitro degradation by diphenylfurans.” Nucleic Acids Res. 2020, in press. DOI: 10.1093/nar/gkaa585
Padroni, G.; Patwardhan, N. N.; Schapira, M.; Hargrove, A. E. “Systematic analysis of the interactions driving small molecule–RNA recognition.” RSC Med Chem. 2020, 11, 802 – 813. DOI:10.1039/D0MD00167H.

Davila-Calderon, Jesse; Patwardhan, Neeraj N.; Chiu, Liang-Yuan; Sugarman, Andrew L.; Cai, Zhengguo; Penutmutchu, Srinivasa R.; Li, Mei-Ling; Brewer, Gary; Hargrove, Amanda E.; Tolbert, Blanton S. “IRES-targeting small molecule inhibits enterovirus 71 replication via allosteric stabilization of a ternary complex.” Nat Comm. 2020, 11, 4775. DOI:10.1038/s41467-020-18594-3
This work was featured on Duke’s Research Blog!
Ganser, Laura R.; Kelly, Megan L.; Patwardhan, Neeraj N.; Hargrove, Amanda E.; Al-Hashimi, Hashim M. “Demonstration that small molecules can bind and stabilize low-abundance short-lived RNA excited conformational states.” J. Mol. Bio. 2020, 432(4), 1297-1304. DOI: 10.1016/j.jmb.2019.12.009
Morgan, B. S.; Sanaba, B. G.; Donlic, A.; Karloff, D. B.; Forte, J. E.; Zhang, Y.; Hargrove, A. E. “R-BIND: An Interactive Database for Exploring and Developing RNA-Targeted Chemical Probes.” ACS Chemical Biology, 2019, 14(12), 2691-2700. DOI: 10.1021/acschembio.9b00631
 Patwardhan, N. N.; Cai, Z.; Umuhire Juru, A.; Hargrove, A. E. “Driving factors in amiloride recognition of HIV RNA targets.” Organic & Biomolecular Chemistry, 2019, 17, 9313-9320. DOI: 10.1039/C9OB01702J
Patwardhan, N. N.; Cai, Z.; Umuhire Juru, A.; Hargrove, A. E. “Driving factors in amiloride recognition of HIV RNA targets.” Organic & Biomolecular Chemistry, 2019, 17, 9313-9320. DOI: 10.1039/C9OB01702J
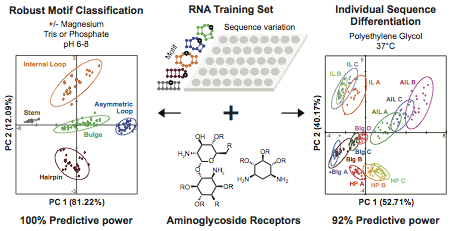
Padroni, G.; Eubanks, C. S.; Hargrove, A. E. “Differentiation and classification of RNA motifs using small molecule-based pattern recognition.” Methods in Enzymology, 2019, 623, 101-130. DOI: 10.1016/bs.mie.2019.05.022
Wicks, S. L.; Hargrove, A. E. “Fluorescent indicator displacement assays to identify and characterize small molecule interactions with RNA.” Methods. 2019, 167, 3-14. DOI: 10.1016/j.ymeth.2019.04.018

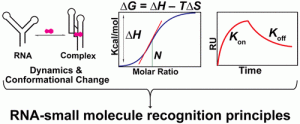
Juru, A. U.; Patwardhan, N. N.; Hargrove, A. E. “Understanding the Contributions of Conformational Changes, Thermodynamics, and Kinetics of RNA–Small Molecule Interactions.” ACS Chem. Biol. 2019, 14(5), 824-838. DOI: 10.1021/acschembio.8b00945
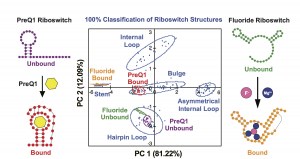
Eubanks, C. S.; Zhao, B.; Patwardhan, N. N.; Thompson, R. D.; Zhang, Q.; Hargrove, A. E. “Visualizing RNA conformational changes via Pattern Recognition of RNA by Small Molecules.” J. Am. Chem. Soc. 2019, 141 (14), 5692-5698. DOI: 10.1021/jacs.8b09665
Eubanks, C. S.; Hargrove, A. E. “RNA Structural Differentiation: Opportunities with Pattern Recognition.” Biochem. 2019, 58, 199-213. DOI: 10.1021/acs.biochem.8b01090
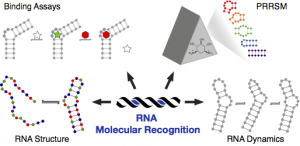
Part of the Future of Biochemistry: The International Issue special issue.
Patwardhan, N. P.; Cai, Z.; Newson, C. N.; Hargrove, A. E. “Fluorescent peptide displacement assay as a general assay for screening small molecule libraries against RNA.” Org. Biomol. Chem. 2019, 17, 1778 – 1786. DOI: 10.1039/c8ob02467g
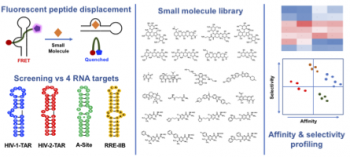
This article is part of themed collections: New Talent and Chemical Biology in OBC.
This work was also highlighted in the Organic and Biomolecular Chemistry blog!
Donlic, A.; Morgan, B. S.; Xu, J. L.; Liu, A.; Roble Jr., C.; Hargrove, A. E. “Discovery of Small Molecule Ligands for MALAT1 by Tuning an RNA-Binding Scaffold.” Angew. Chem. Int. Ed. 2018, 40, 13242-13247. DOI: 10.1002/anie.201808823

Chen, G. and Edwards, N: F1000Prime Recommendation of Evaluation [Donlic A et al., Angew Chem Int Ed Engl 2018 57(40):13242-13247]. In F1000Prime, 21 Nov 2018; 10.3410/f.733852452.793552881
Morgan, B. S.; Forte, J. E.; Hargrove, A.E. “Insights into the Development of Chemical Probes for RNA.” Nucleic Acids Res. 2018, 16, 8025-8037. DOI: 10.1093/nar/gky718

Chaires J: F1000Prime Recommendation of Evaluation [Morgan BS et al., Nucleic Acids Res 2018 46(16):8025-8037]. In F1000Prime, 04 Sep 2018; 10.3410/f.733808492.793550119
Donlic, A.; Hargrove, A. E. “Targeting RNA in Mammalian Systems with Small Molecules.” WIREs RNA 2018, 9:(4) e1477. DOI: 10.1002/wrna.1477
This article was featured in Advanced Science News!
Eubanks, C. S.; Hargrove, A. E. “Sensing the Impact of Environment on Small Molecule Differentiation of RNA Sequences.” Chem. Comm. 2017, 53, 13363-13366. DOI: 10.1039/C7CC07157D
This article is part of the themed collection: Chemosensors and Molecular Logic
Morgan, B. S.; Forte, J. E.; Culver, R. N.; Zhang, Y.; Hargrove, A. E. “Discovery of Key Physicochemical, Structural, and Spatial Properties of RNA-targeted Bioactive Ligands.” Angew. Chem. Int. Ed. 2017, 56, 13498-13502. DOI: 10.1002/anie.201707641

This work was featured on Duke’s Research Blog!
Patwardhan, N. N.; Ganser, L. R.; Kapral, G. J.; Eubanks, C. S.; Lee, J.; Sathyamoorthy, B.; Al-Hashimi, H. M.; Hargrove, A. E. “Amiloride as a new RNA-binding scaffold with activity against HIV-1 TAR.” Med. Chem. Comm. 2017, 8, 1022-1036. DOI: 10.1039/C6MD00729E

This article is part of the themed collection: 2017 Hot Articles in MedChemComm
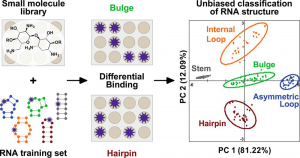
Eubanks, C. S.; Forte, J. E.; Kapral, G. J.; Hargrove, A. E. “Small Molecule-Based Pattern Recognition to Classify RNA Structure.” J. Am. Chem. Soc. 2017, 139, 409-416. DOI: 10.1021/jacs.6b11087.
Arora P and Barros S: F1000Prime Recommendation of Evaluation [Eubanks CS et al., J Am Chem Soc 2017 139(1):409-416]. In F1000Prime, 07 Jun 2017; 10.3410/f.727137423.793532776
Bujnicki J and Almeida C: F1000Prime Recommendation of Evaluation [Eubanks CS et al., J Am Chem Soc 2017 139(1):409-416]. In F1000Prime, 21 Jun 2017; 10.3410/f.727137423.793533363
McFadden, E. J.; Hargrove, A. E. “Biochemical Methods To Investigate lncRNA and the Influence of lncRNA:Protein Complexes on Chromatin.” Biochem. 2016, 55, 1615-1630. DOI: 10.1021/acs.biochem.5b01141.

Morgan, B. S.; Hargrove, A. E., Chapter 7 Synthetic Receptors for Oligonucleotides and Nucleic Acids. In Synthetic Receptors for Biomolecules: Design Principles and Applications, The Royal Society of Chemistry: 2015; pp 253-325. DOI: 10.1039/9781782622062-00253

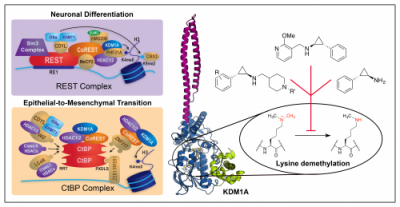
Burg, J. M.; Link, J. E.; Morgan, B. S.; Heller, F. J.; Hargrove, A. E.; McCafferty, D. G. “KDM1 Class Flavin-Dependent Protein Lysine Demethylases.” Biopolymers 2015, 104, 213-246. DOI: 10.1002/bip.22643
Previous Work
Hargrove, A. E.; Martinez, T. F.; Hare, A. A.; Kurmis, A. A.; Philips, J. W.; Sud, S.; Pienta, K. J.; Dervan, P. B. “Tumor Repression of VCaP Xenografts by a Pyrrole-Imidazole Polyamide.”PLoS One, 2015, 10, 11.
Nickols, N. G.; Szablowski, J. O.; Hargrove, A. E.; Li, B. C.; Raskatov, J. A.; Dervan, P. B. “Activity of a Py-Im polyamide targeted to the estrogen response element.” Mol. Cancer Ther. 2013, 12, 675-684.
Raskatov, J. A.; Nickols, N. G.; Hargrove, A. E.; Marinov, G.; Wold, B.; Dervan, P. B. “Gene Expression Changes in a Tumor Xenograft by a Py-Im Polyamide.” Proceedings of the National Academy of Sciences. 2012, 109, 16041-16045.
Hargrove, A. E.; Raskatov, J. A.; Meier, J. L.; Montgomery, D. C.; Dervan, P. B. “Characterization and solubilization of pyrrole-imidazole polyamide aggregates. J. Med. Chem. 2012, 55, 5425-5432.
Raskatov, J. A.; Hargrove, A. E.; So, A. Y.; Dervan, P. B. “Pharmacokinetics of Py-Im Polyamides Depend on Architecture: Cyclic versus Linear.” J. Am. Chem. Soc. 2012, 134(18), 7995-7999.
Hargrove, A. E.; Nieto, S.; Zhang, T.; Sessler, J. L.; Anslyn, E. V. “Artificial receptors for the recognition of phosphorylated molecules.” Chem. Rev. 2011, 111(11), 6603-6782.
Hargrove, A. E.; Ellington, A. D.; Anslyn, E. V.; Sessler, J. L. “Chemical functionalization of oligodeoxynucleotides with multiple boronic acids for the polyvalent binding of saccharides.” Bioconjugate Chem. 2011, 22, 388-396.
Hargrove, A. E.; Reyes, R. N.; Riddington, I.; Anslyn, E. V.; Sessler, J. L. “Boronic acid porphyrin receptor for ginsenoside sensing.” Org. Lett. 2010, 12(21), 4804-4807.
Hargrove, A. E.; Zhong, Z.; Shabbir, S. H.; Sessler, J. L., Anslyn, E. V. “Algorithms for the determination of binding constants and enantiomeric excess in complex host:guest equilibria using optical measurements.” New J. Chem. 2010, 34, 348-354
Collins, B. E.; Sorey, S.; Hargrove, A. E.; Shabbir, S. H.; Lynch, V. M.; Anslyn, E. V. “Probing intramolecular B-N interactions in protic media.” J. Org. Chem. 2009, 74(11), 4055 – 4066.
Pikulski, M.; Hargrove, A. E.; Shabbir, S. H.; Anslyn, E. V.; Brodbelt, J. S. “Sequencing and characterization of oligosaccharides using infrared multiphoton dissociation and boronic acid derivatization in a quadrupole ion trap.” J. Am. Soc. Mass Spectr. 2007, 18, 2094-2106.
Spence, J. D.; Hargrove, A. E.; Crampton, H. L.; Thomas, D. W. “Porphyrenediynes: synthesis and cyclization of meso-enediynylporphyrins.” Tetrahedron Lett. 2007, 48(4), 725-728.
Sessler, J. L.; Lee, J. T.; Ou, Z.; Köhler, T.; Hargrove, A. E.; Cho, W; Lynch, V; Kadish, K. M. “Chemical and electrochemical oxidation of N-alkyl cyclo[n]pyrroles.” J. Porphyr. Phthalocya. 2006, 10, 1329-1336.